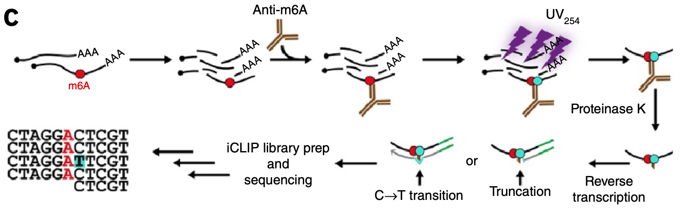
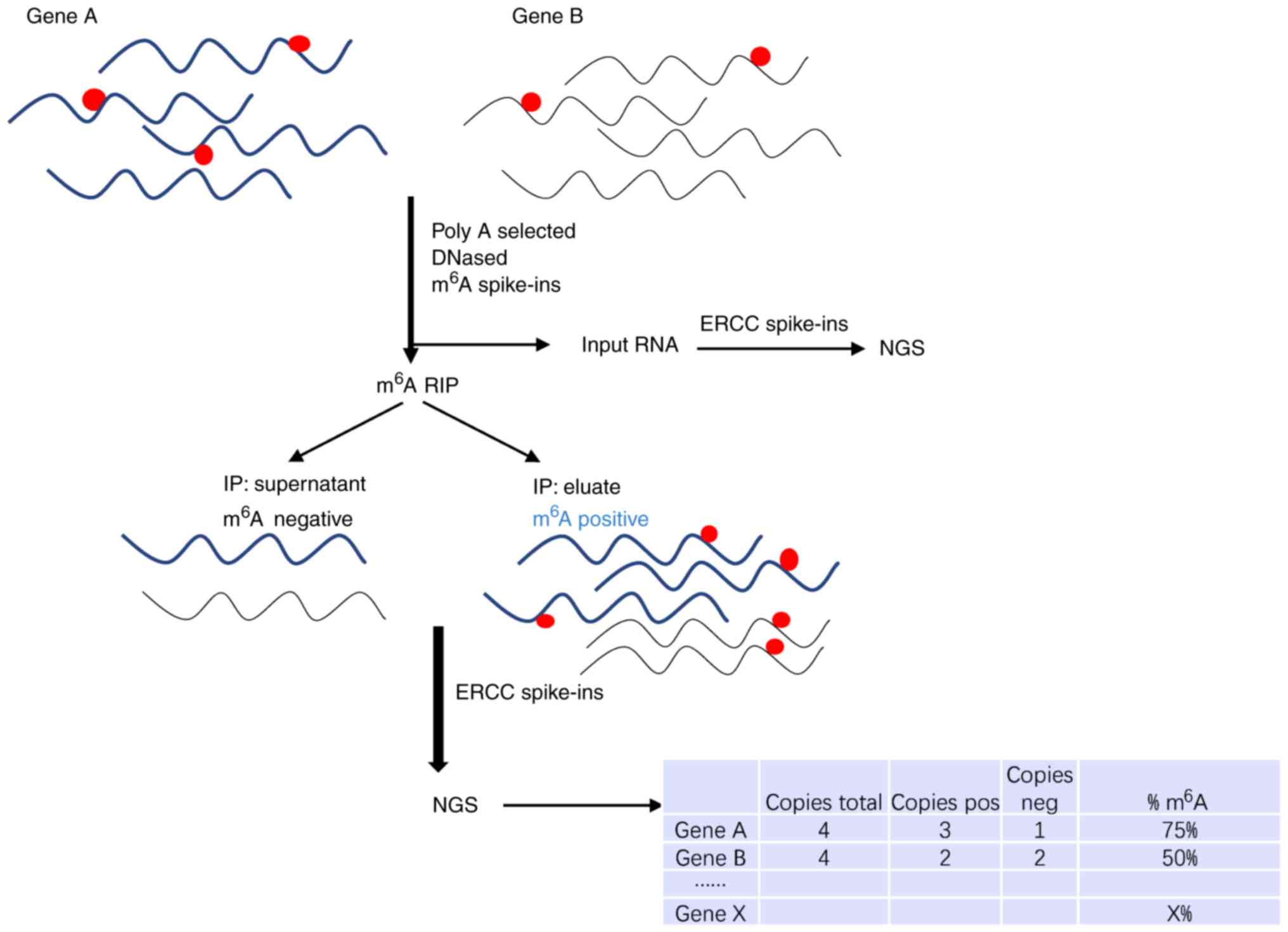
Single-nucleotide resolution achieved by m 6 A-CLIP/immunoprecipitation... | Download Scientific Diagram

Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses | Scientific Reports

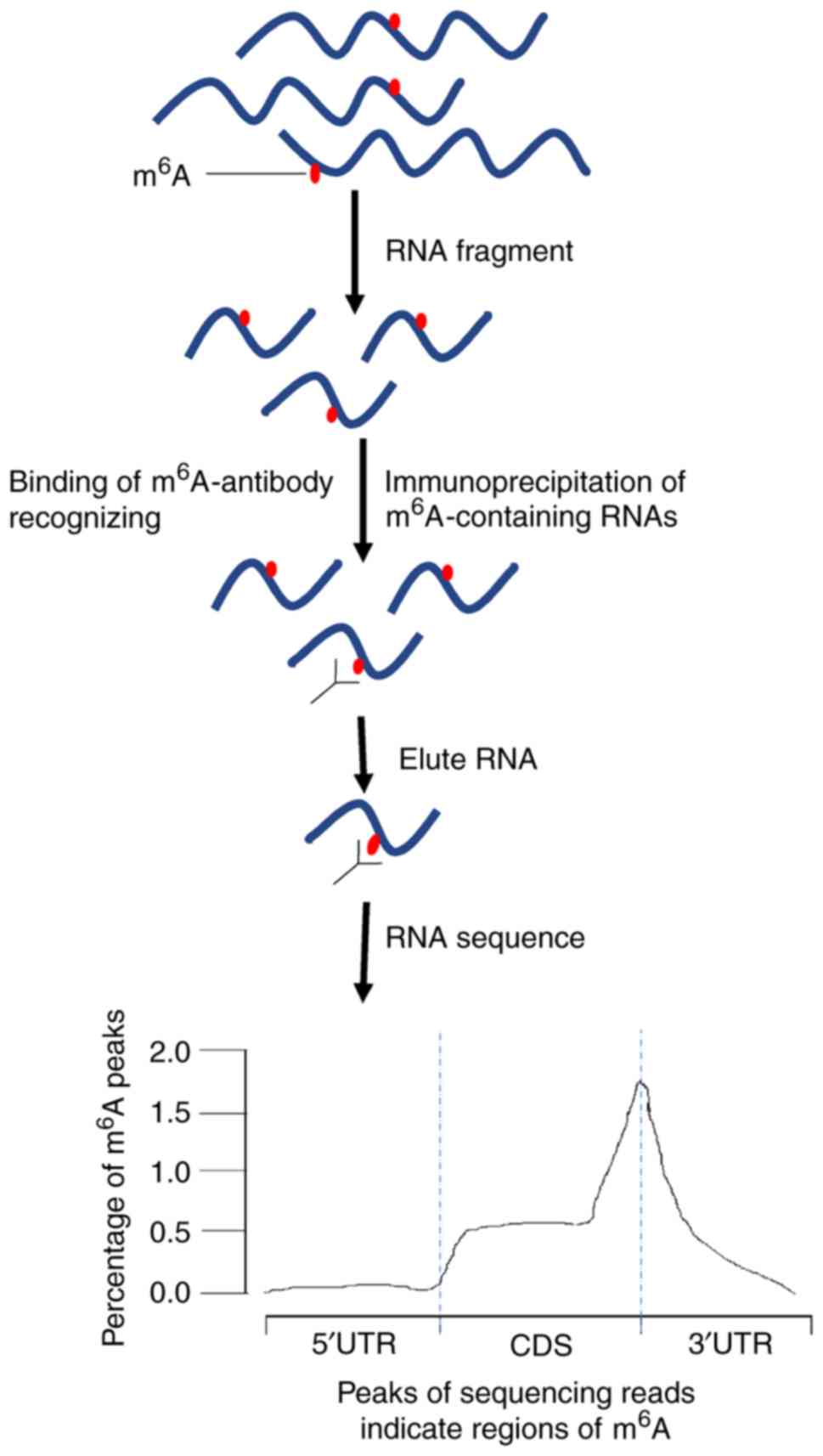
High‐Resolution N6‐Methyladenosine (m6A) Map Using Photo‐Crosslinking‐Assisted m6A Sequencing - Chen - 2015 - Angewandte Chemie International Edition - Wiley Online Library
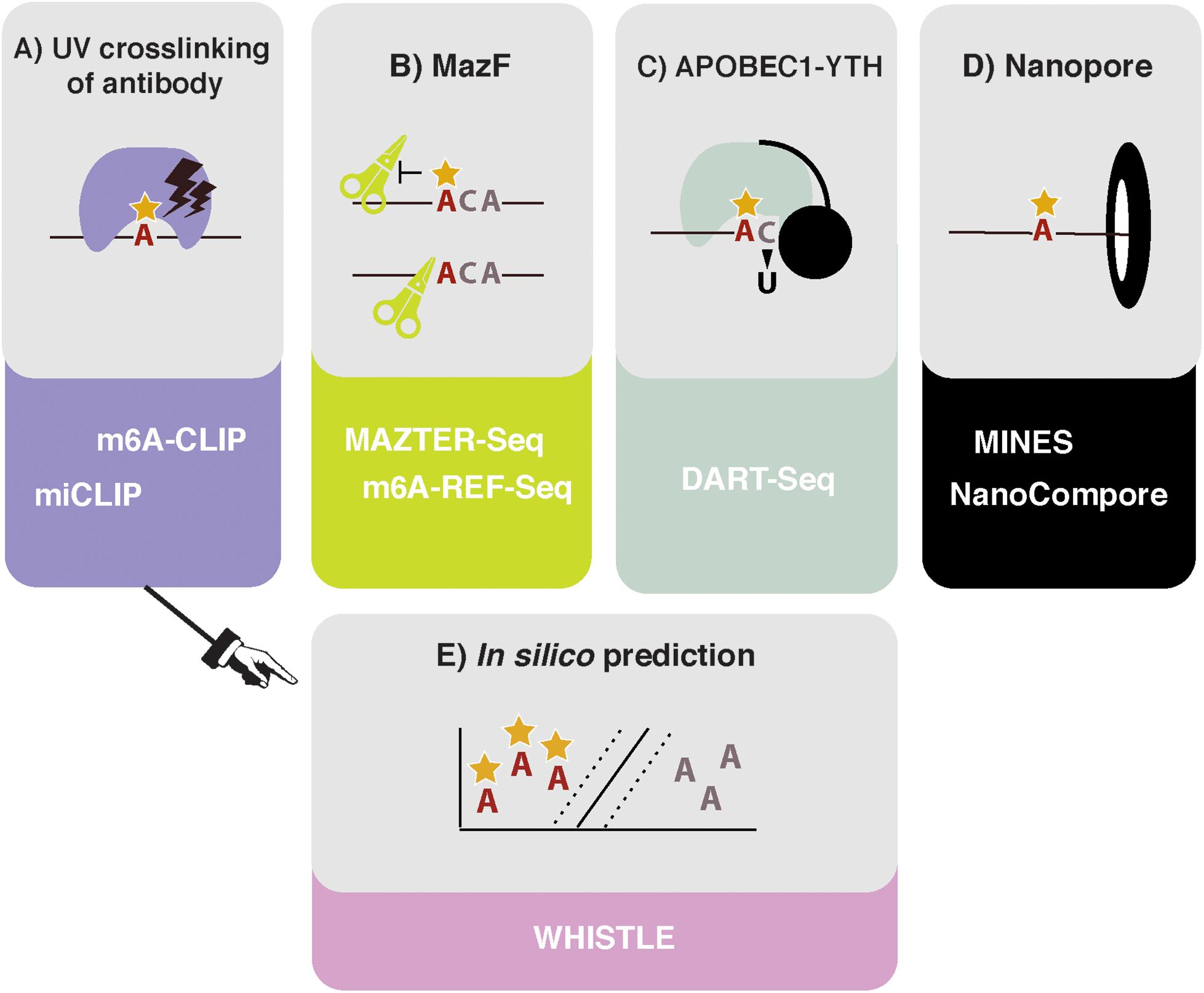
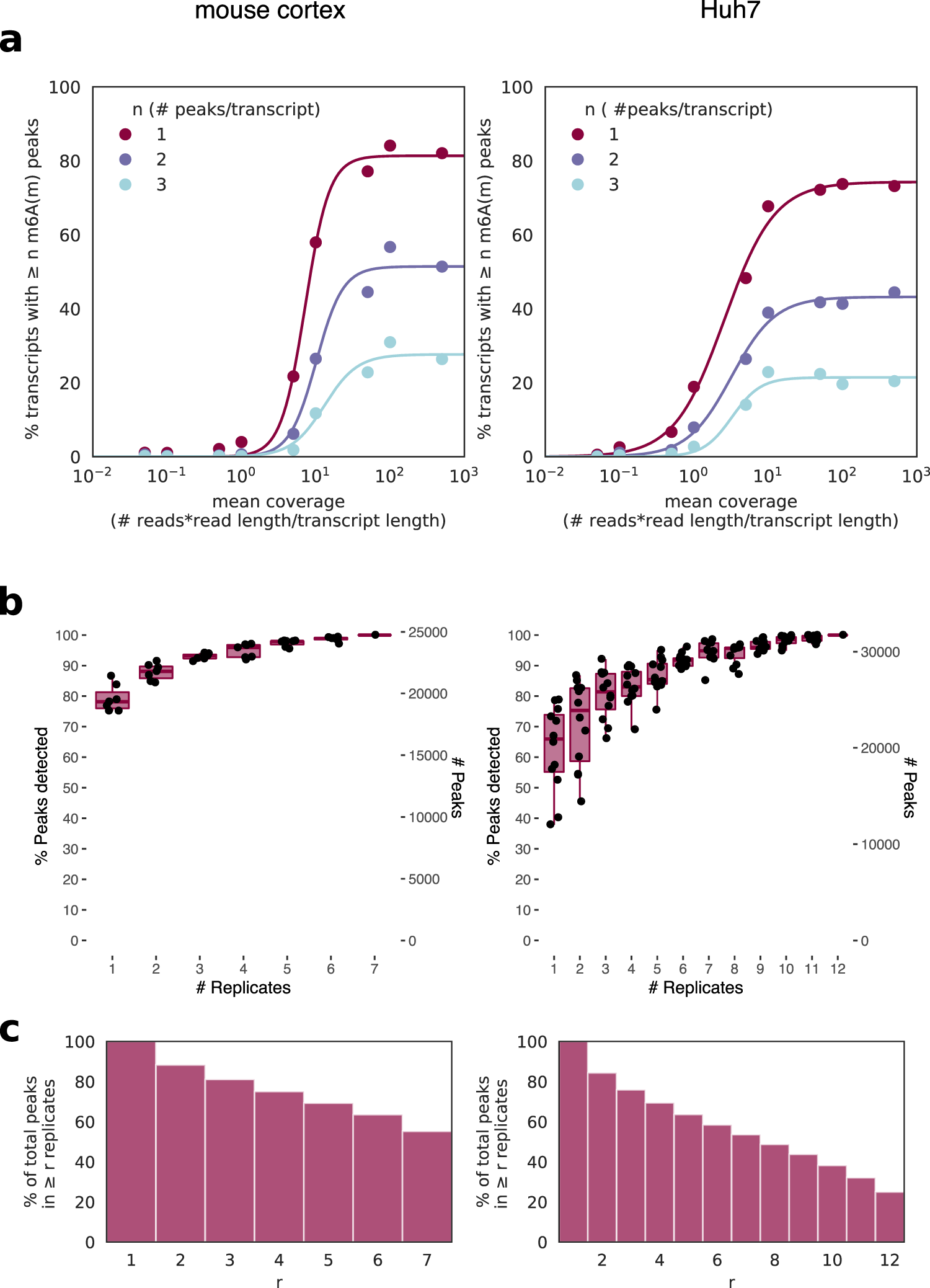
Frontiers | How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets

m6A reader Pho92 is recruited co-transcriptionally and couples translation efficacy to mRNA decay to promote meiotic fitness in yeast | bioRxiv

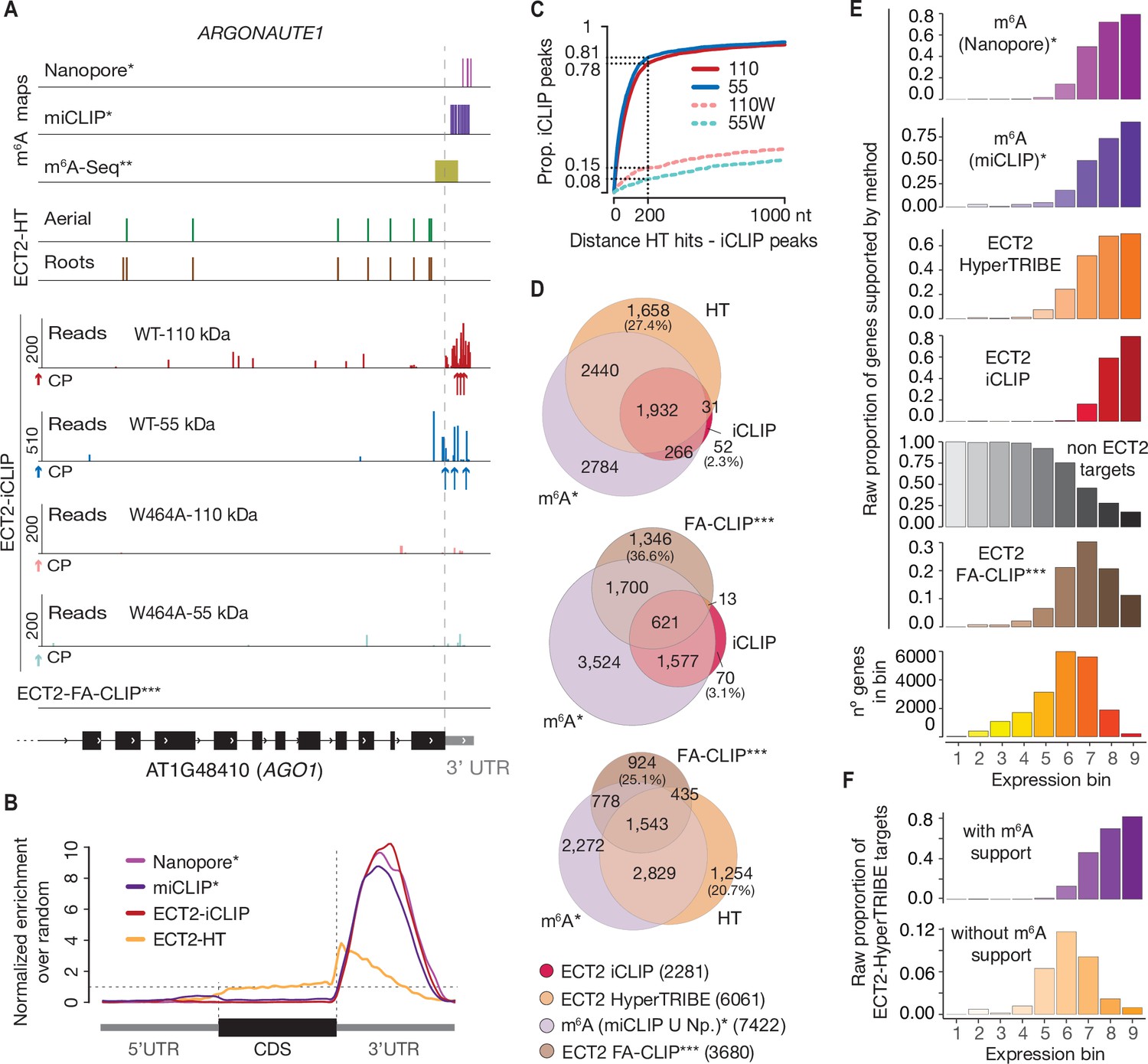
Acute depletion of METTL3 identifies a role for N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome | bioRxiv

Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine | Nature Chemical Biology

N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. – 武汉生命之美科技有限公司

Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning | bioRxiv
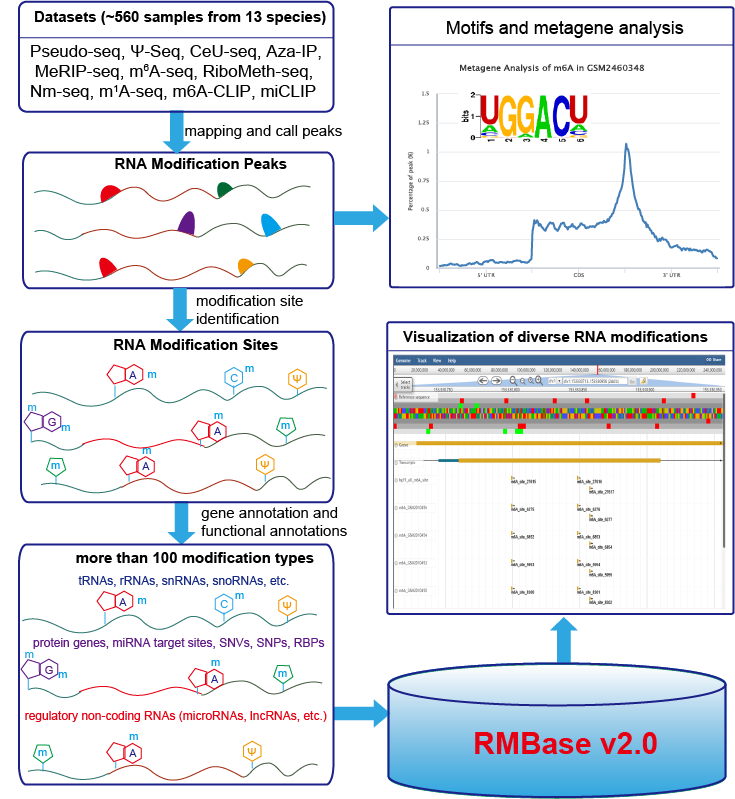
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).